* What can do Cov-HiPathia for you
* How to use Cov-HiPathia:
* Tools:
* Worked examples:
* For further learning:
* What can do Cov-HiPathia for you
* How to use Cov-HiPathia:
* Tools:
* Worked examples:
* For further learning:
HiPathia allows us to compute a signal value through a chosen signaling pathways and for each one of our data samples. Therefore, it allows us to compare the signal value between different conditions. We can:
The tool can be accessed from the main menu bar, by clicking on the Differential signaling button, see Workflow for further information.
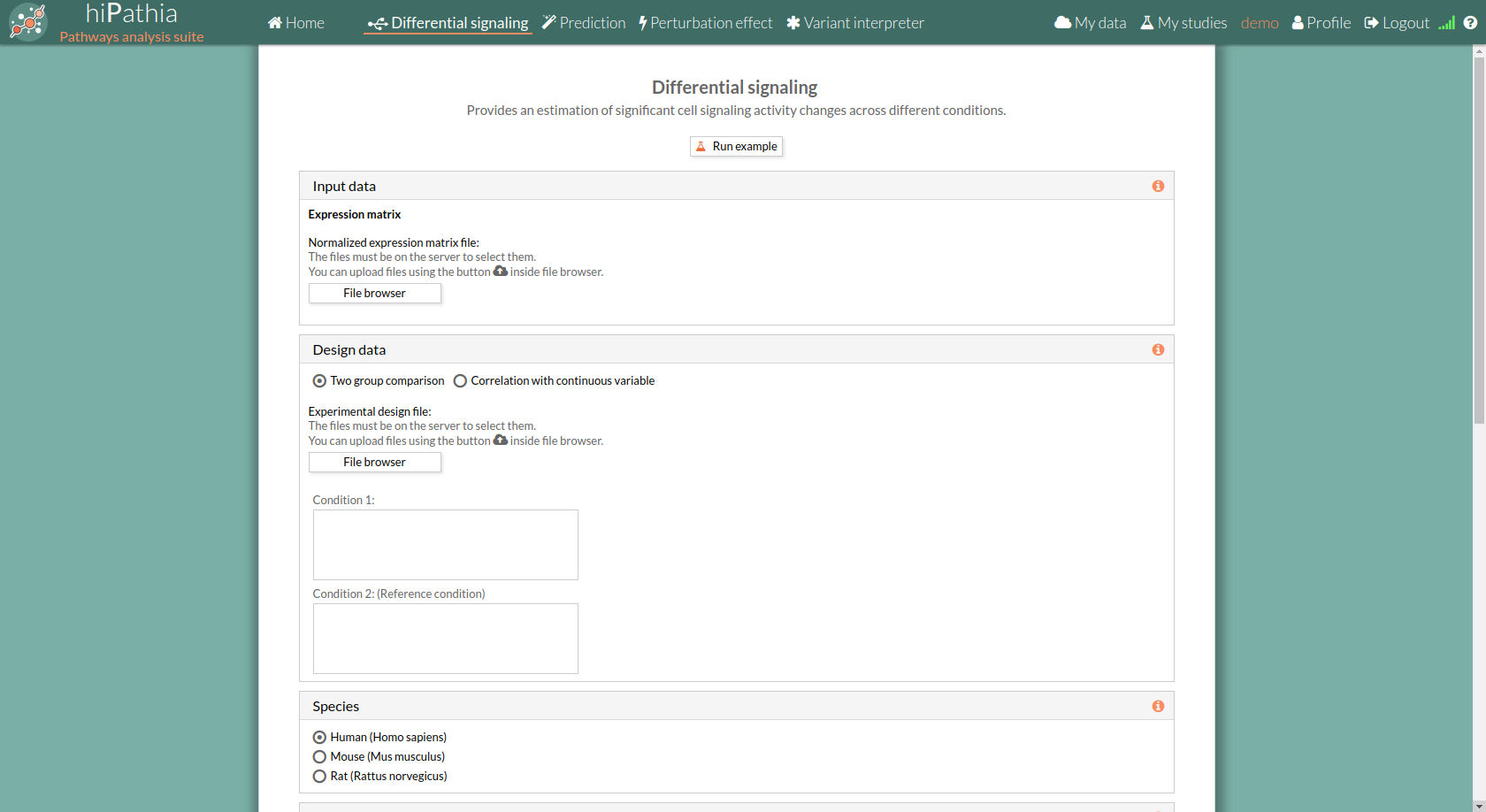
The main page of the tool is its filling in form. This form includes all the information and parameters that the tool needs to process a Differential signaling study. The form is divided in different panels:
In the input data panel, we must introduce the expression data.
 The expression data is a gene expression matrix provided by ourselves (see how to upload files in Upload your data).
The expression data is a gene expression matrix provided by ourselves (see how to upload files in Upload your data).
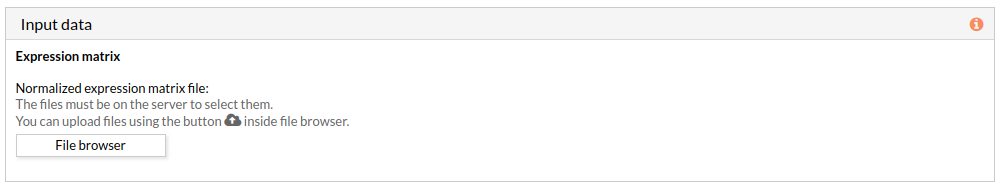
When we select a gene expression file, the number of samples of this matrix will appear under the “file browser” button as shown below.
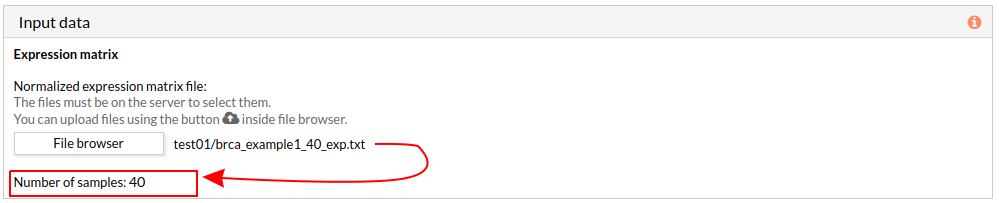
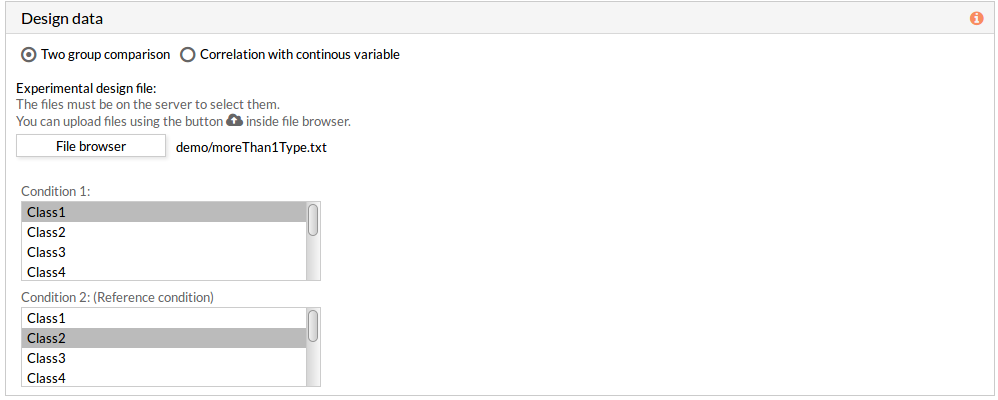
The design data panel allows you to choose the kind of experiment you want to perform. You can choose between two kinds of experimental design:
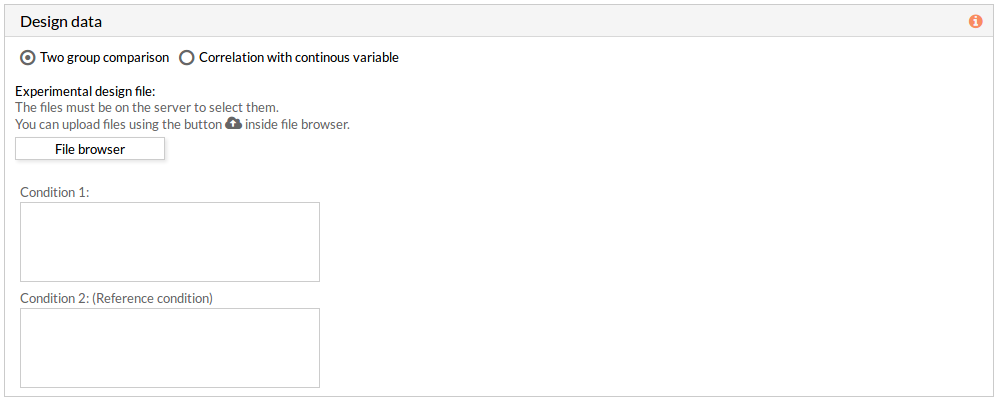 If the experimental design file contains more than two classes then you have to select the appropriate class for each condition.
If the experimental design file contains more than two classes then you have to select the appropriate class for each condition.
Note: the condition 2 will be taken as a reference condition.
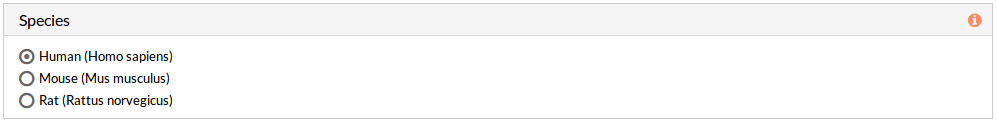
Here we must choose the species of our experiment. You can choose among:
This panel includes further parameters necessary to run a study.
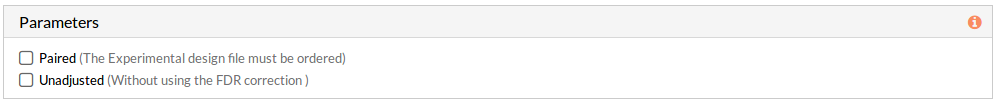
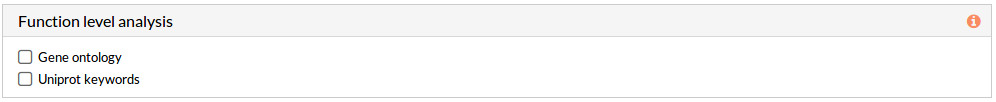
If any of these checkboxes is selected, a differential functional activity analysis is performed.
From a given gene expression profile, we estimate an effector proteins activation profiles and then we annotate each effector proteins using Gene ontology and Uniprot keywords annotation.
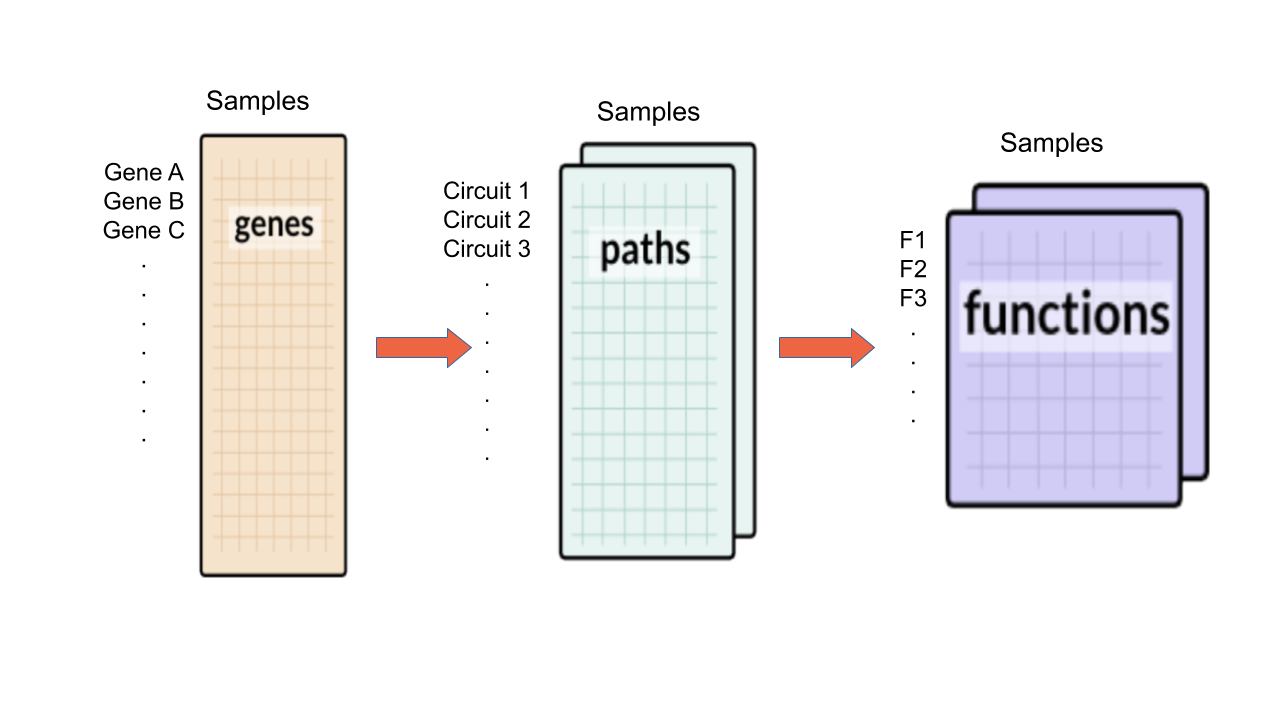
This panel includes the list of all available pathways in HiPathia. We can select the pathways with which the analysis will be performed.
HiPathia retrieves pathway information from KEGG database. KEGG pathway database is a collection of manually drawn pathway maps representing the knowledge on the molecular interaction, reaction and relation networks.
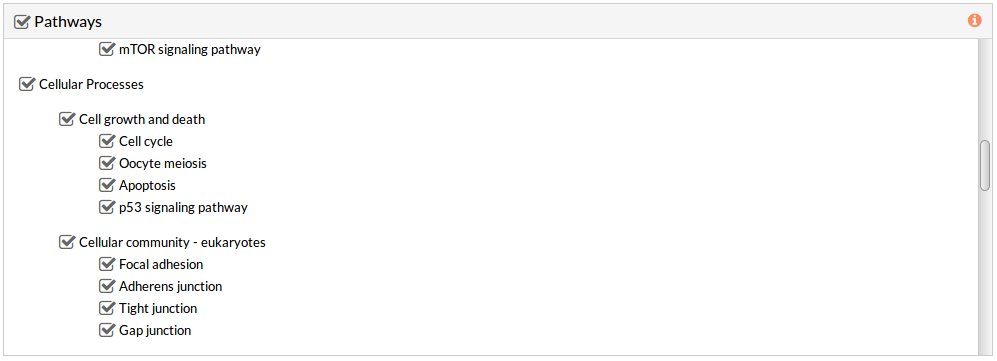 By default all available pathways are selected.
By default all available pathways are selected.
Note: At least one pathway has to be selected.
This panel includes some parameters in order to identify and save our study.
Once the form has been filled in, press the Run analysis button to launch the study.
 Your study will be listed in the My studies panel, and a panel called Browse my studies will appear showing all your studies and their state. the new study will appear with a queued state then running state. If everything goes well, the state will be done after few minutes(depending on the inputs data and the availability of server).
All study states are:
Your study will be listed in the My studies panel, and a panel called Browse my studies will appear showing all your studies and their state. the new study will appear with a queued state then running state. If everything goes well, the state will be done after few minutes(depending on the inputs data and the availability of server).
All study states are:

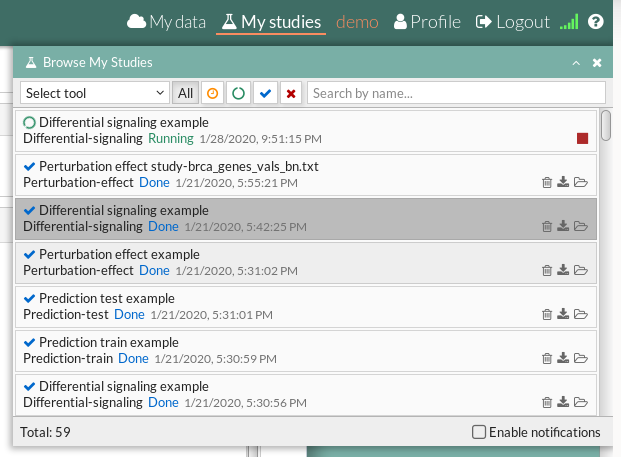
When an analysis is finished at Hipathia web, the report of such analysis will be available in “My studies” section once the study is done.
The results page of the Differential signaling tool includes different output results. You can download any table or image showed in the report page by clicking on the name right before it.
You can also download the circuit activity values and function matrices by clicking on Circuit values, GO terms values and Uniprot keywords values respectively.
Furthermore, the report can be downloaded in order to visualize it locally.
 The downloaded RAR file has to be extracted then you can open the “index.html” file using a web browser like Firefox web browser, some configuration may be needed for chrome or other web browsers: installation of web server( for example: Wamp or XAMPP) then open the report from the navigator using http://localhost/Path/to/my/downloaded_report/)
The downloaded RAR file has to be extracted then you can open the “index.html” file using a web browser like Firefox web browser, some configuration may be needed for chrome or other web browsers: installation of web server( for example: Wamp or XAMPP) then open the report from the navigator using http://localhost/Path/to/my/downloaded_report/)
The results are divided in different panels:
Here you can find the information about the selected study.

Here you can visualize the parameters with which the current study was launched.

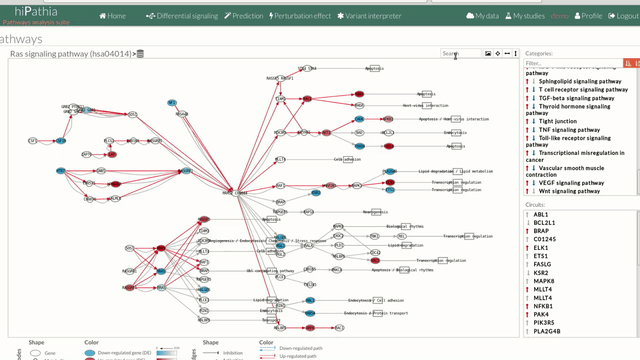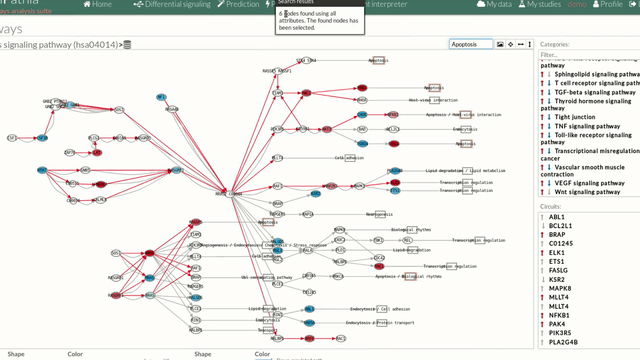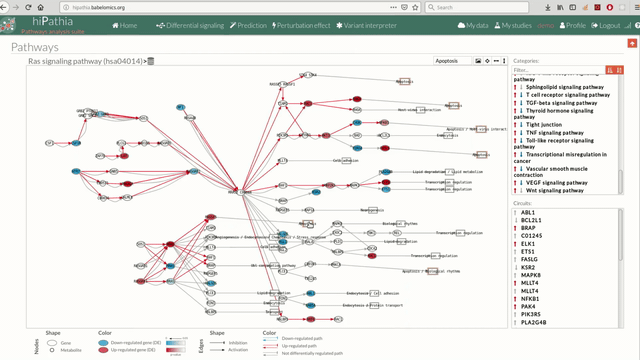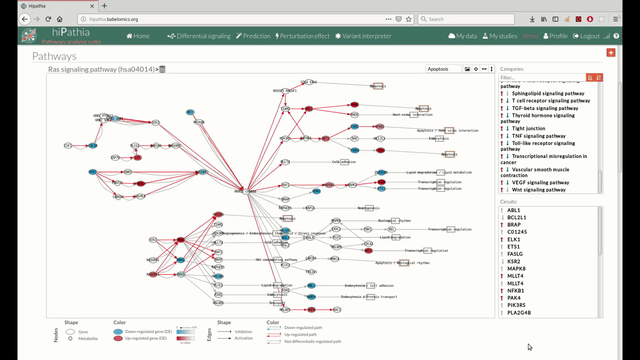
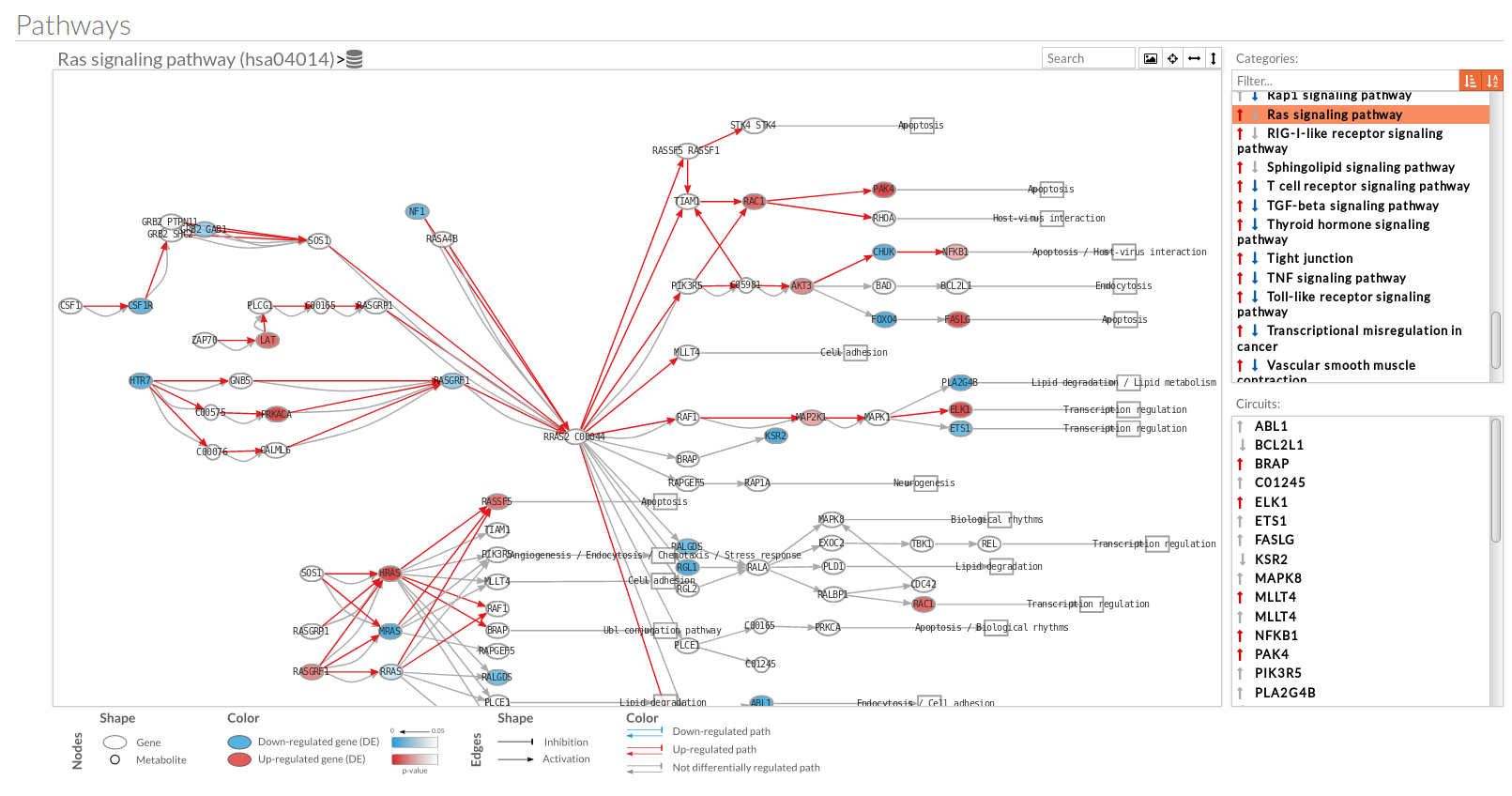 A pathway viewer has been developed in order to make easier the visualization of the comparison results. it shows a graphical representation of the comparison on the activation levels of the “Effector circuit” between the two groups. The meaning of the different symbols that can be found are in the legend below.
A pathway viewer has been developed in order to make easier the visualization of the comparison results. it shows a graphical representation of the comparison on the activation levels of the “Effector circuit” between the two groups. The meaning of the different symbols that can be found are in the legend below.
 The information summarized in the pathway viewer includes:
The information summarized in the pathway viewer includes:
Node's shape:
Node's color:
Edge's shape: Edges represent either activations or inhibitions.
Edge's color:
The pathway viewer has four principal parts in order to facilitate the visualization of the results:
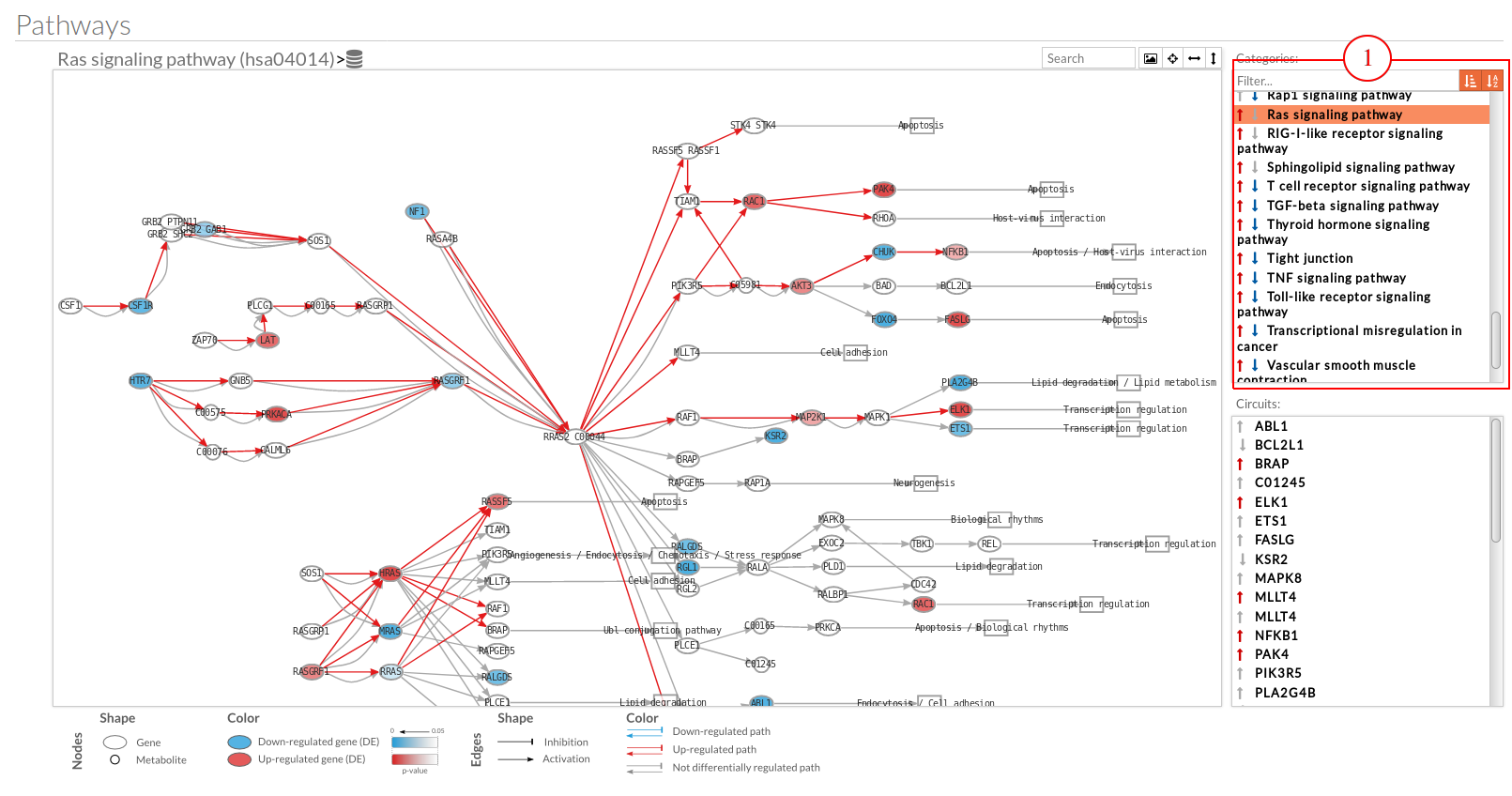
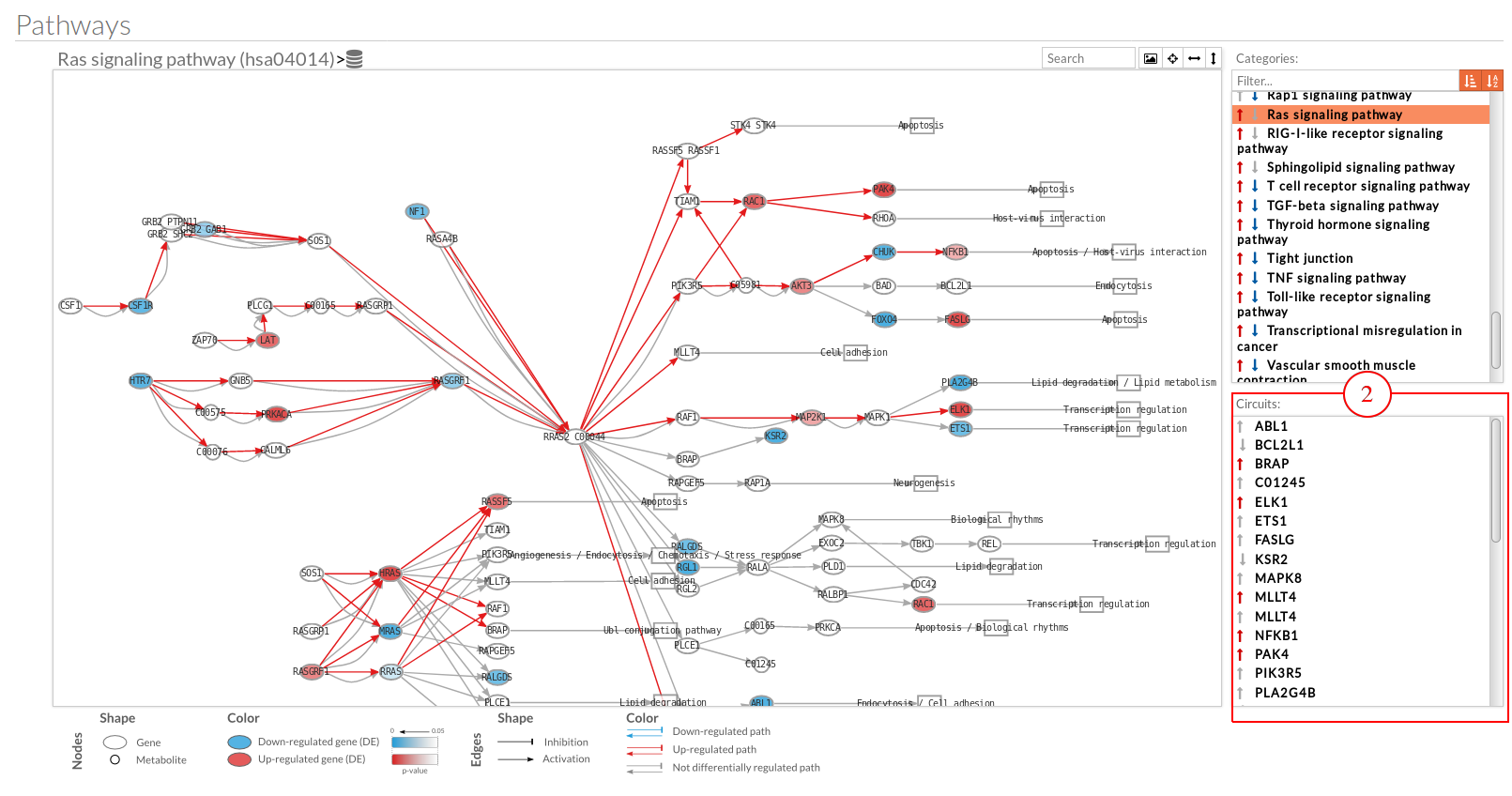
 you can see the original source of this pathway.
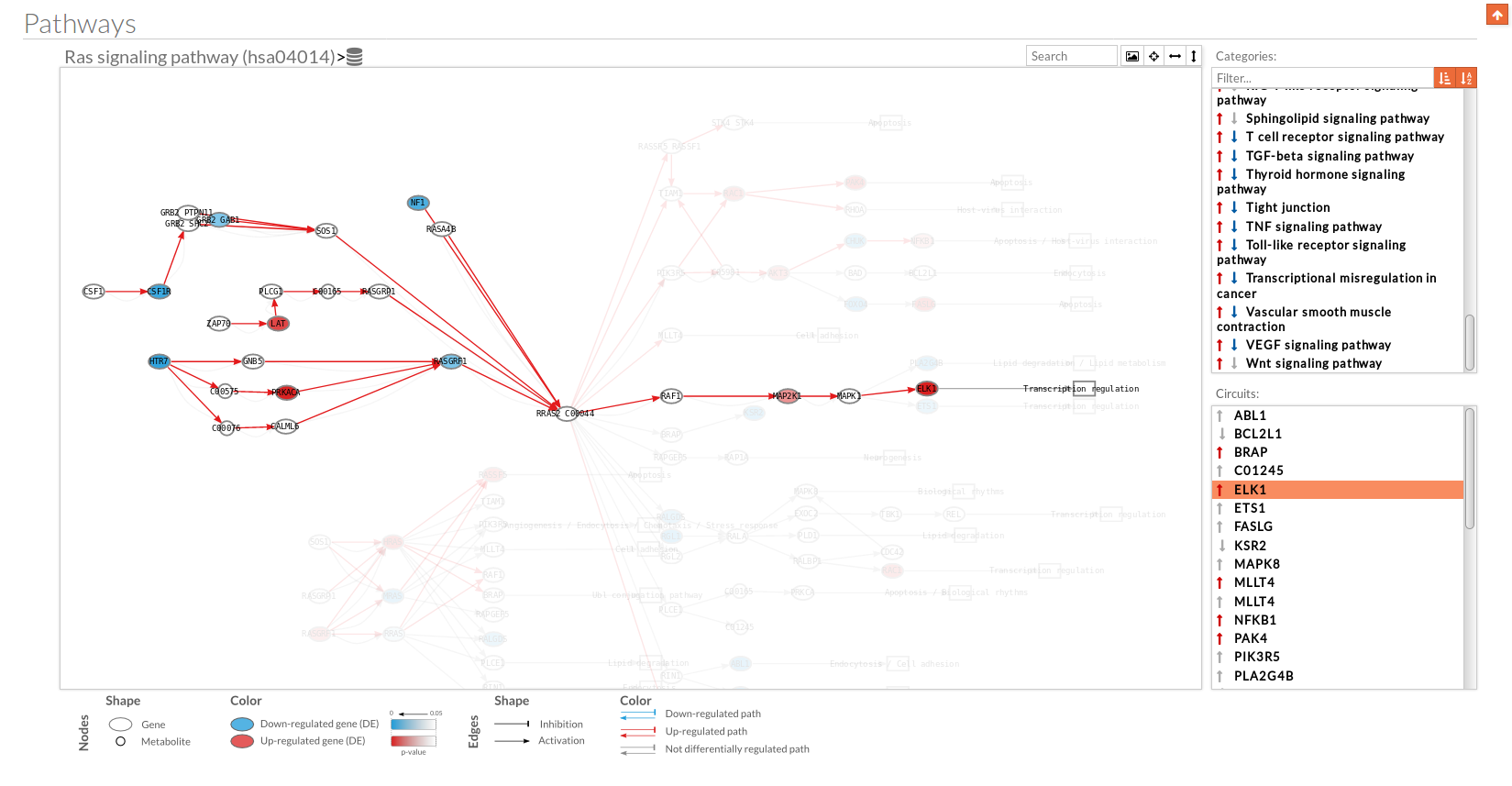
you can see the original source of this pathway. Allow to search specific genes, proteins or functions.
Allow to search specific genes, proteins or functions.
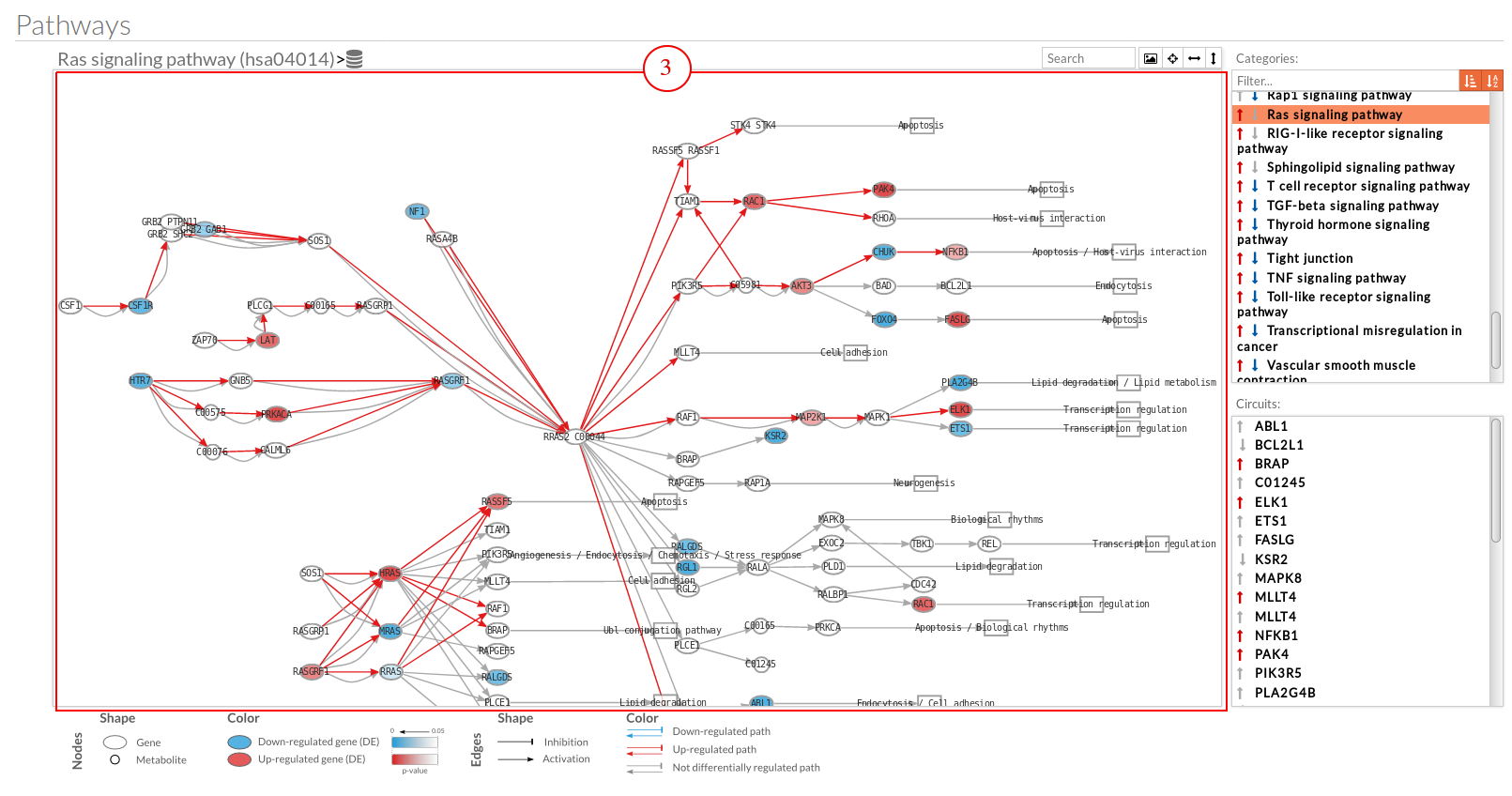
 Export a SVG image of viewed objects (the whole pathway or just the selected effector circuit)
Export a SVG image of viewed objects (the whole pathway or just the selected effector circuit) Center the selected pathway
Center the selected pathway Width adjust
Width adjust  Height adjust
Height adjust Here you can find additional tables and plots:

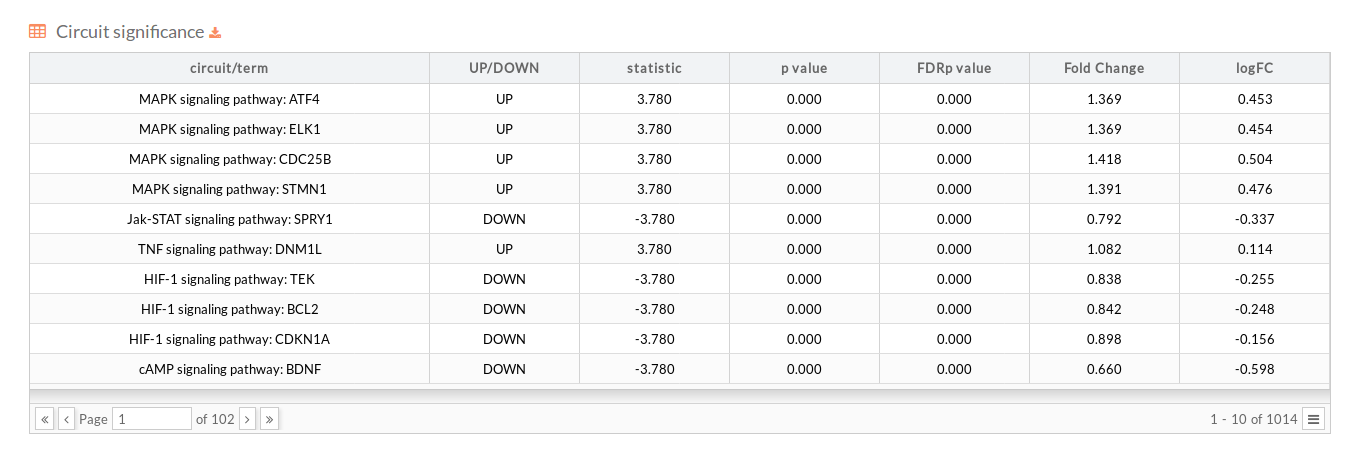
For each of the functional analysis selected to be performed, the same descriptive statistics as in the Path values case are shown: