* What can do Cov-HiPathia for you
* How to use Cov-HiPathia:
* Tools:
* Worked examples:
* For further learning:
* What can do Cov-HiPathia for you
* How to use Cov-HiPathia:
* Tools:
* Worked examples:
* For further learning:
The aim of the Prediction tool is to take advantage of the signalling circuit activities to distinguish between phenotypes. Depending on the study design, we can perform a two group comparison or a correlation with a continuous variable.
HiPathia Prediction uses the signalling value of mechanism-based biomarkers to compute a SVM prediction model with cross-validation. Moreover, previously obtained models could be used to predict the phenotype of new samples.
The tool can be accessed from the main menu bar, by clicking on the Prediction button, see Workflow for further information.
The main page of a the tool is its filling in form. This form includes all the information and parameters that the tool needs to process a study. The form is divided in different panels:
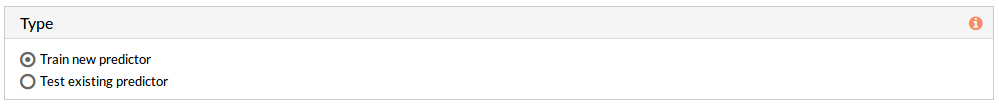
The type panel allows you to choose the kind of prediction analysis you want to perform. You can choose between two options:
In the input data panel, we must introduce the expression data.
 The expression data has to be:
The expression data has to be:
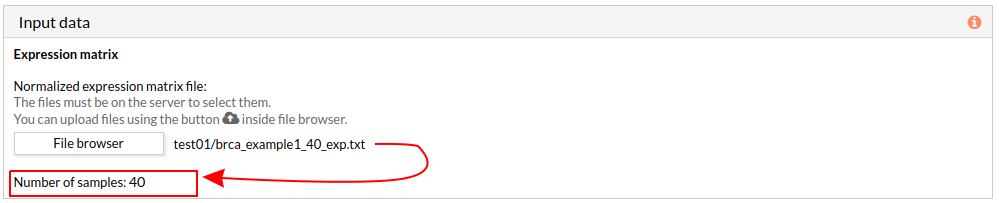
When we select a gene expression file, the number of samples of this matrix will appear under the “file browser” button as shown below.
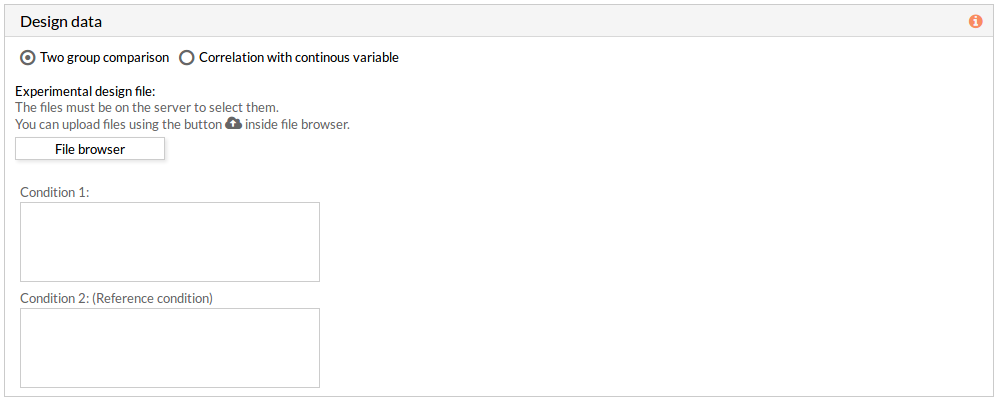
The design data panel allows you to choose the kind of experiment you want to perform. You can choose between two kinds of experimental design:
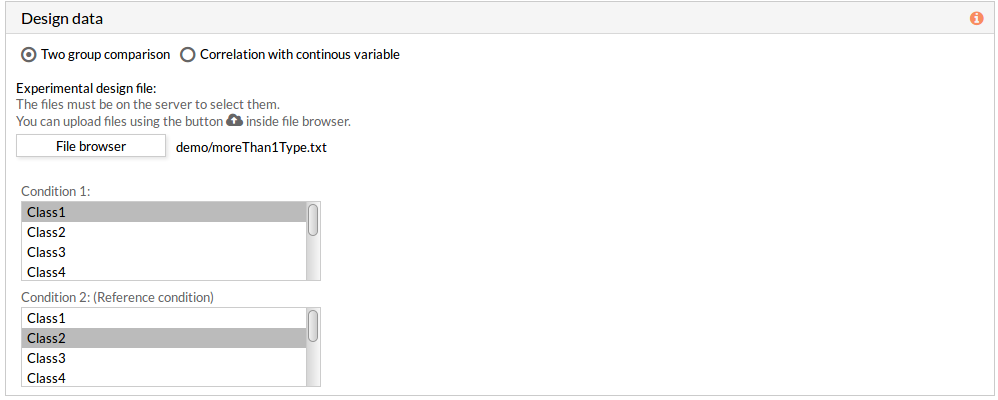
 If the experimental design file contains more than two classes then you have to select the appropriate class for each condition.
If the experimental design file contains more than two classes then you have to select the appropriate class for each condition.
Note: the condition 2 will be taken as a reference condition.
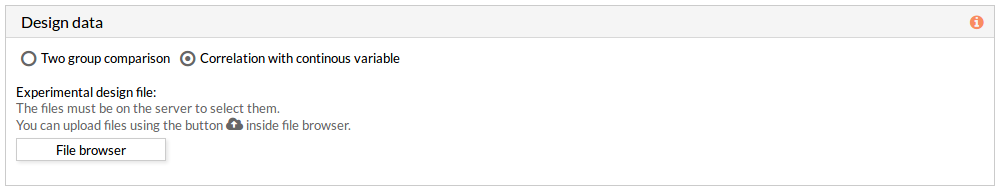
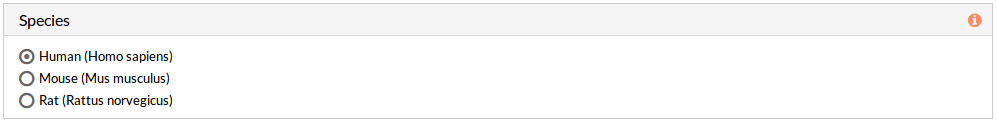
Here we must choose the species of our experiment. You can choose among:
This panel includes further parameters necessary to run an analysis.
Filter circuits: Check to obtain the circuits that best differentiate your phenotype. This option is only available from Prediction tool.

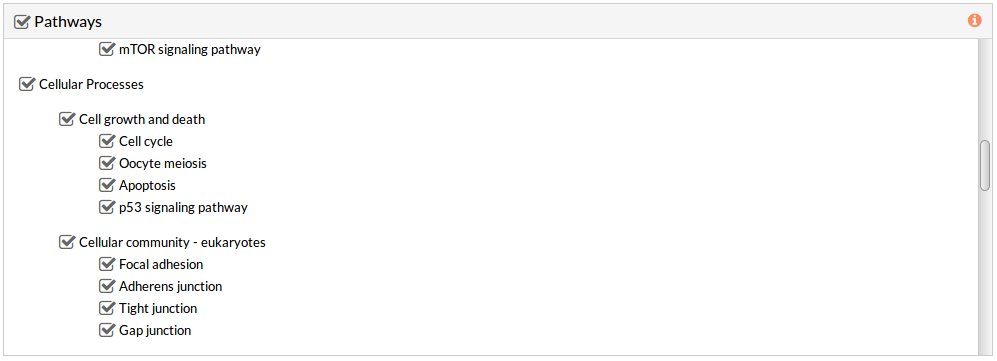
This panel includes the list of all available pathways in HiPathia. We can select the pathways with which the analysis will be performed.
HiPathia retrieves pathway information from KEGG database. KEGG pathway database is a collection of manually drawn pathway maps representing the knowledge on the molecular interaction, reaction and relation networks.
 By default all available pathways are selected.
By default all available pathways are selected.
Note: At least one pathway has to be selected.
This panel includes some parameters in order to identify and save our study.
Once the form has been filled in, press the Run analysis button to launch the study.
 Your study will be listed in the My studies panel, and a panel called Browse my studies will appear showing all your studies and their state. the new study will appear with a queued state then running state. If everything goes well, the state will be done after few minutes(depending on the inputs data and the availability of server).
All study states are:
Your study will be listed in the My studies panel, and a panel called Browse my studies will appear showing all your studies and their state. the new study will appear with a queued state then running state. If everything goes well, the state will be done after few minutes(depending on the inputs data and the availability of server).
All study states are:


The report page of the Prediction tool includes different output results. You can download any table or image showed in the results page by clicking on the name right before it. You can also download the pathway and function matrices by clicking on Circuit values.
The results are divided in different panels:
Here you can find the information about the selected study.

Here you can visualize the parameters with which the current study was launched.

You can download the matrix of circuit activity values by clicking on circuit values.
 This matrix file indicates for each “effector circuit” the level of activation calculated using Hipathia method for each sample.
This matrix file indicates for each “effector circuit” the level of activation calculated using Hipathia method for each sample.
Here you can visualize the results from the prediction analysis.

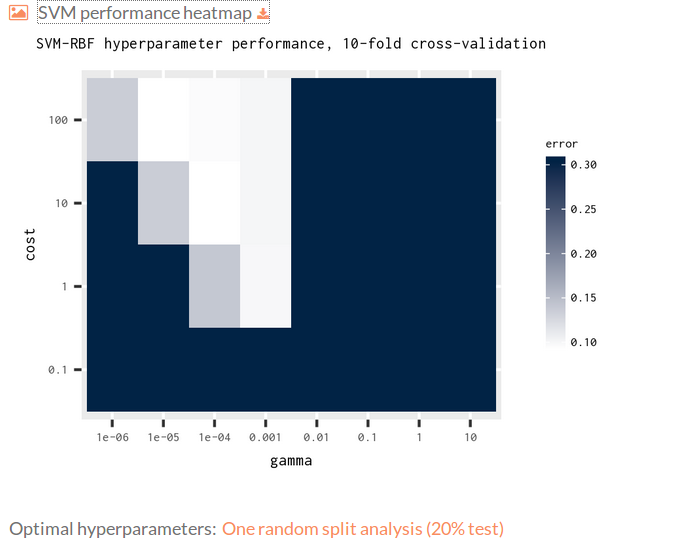
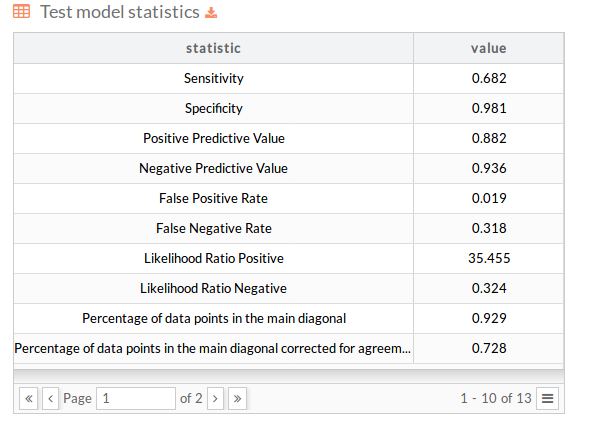
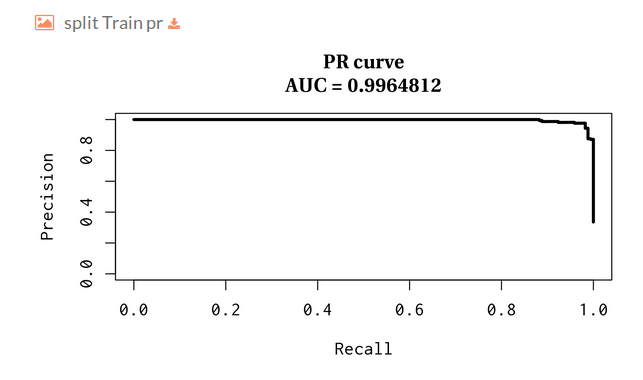
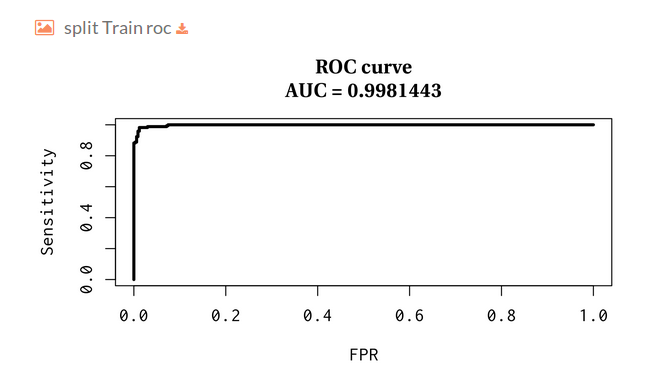
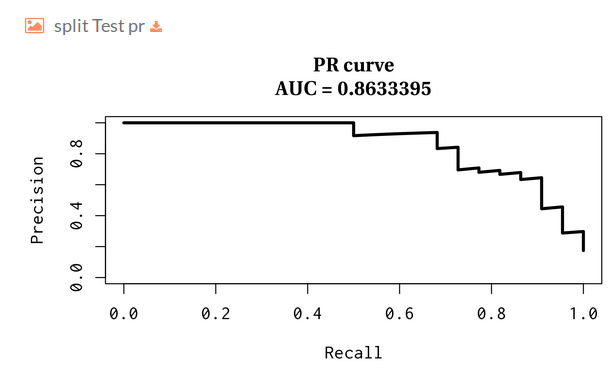
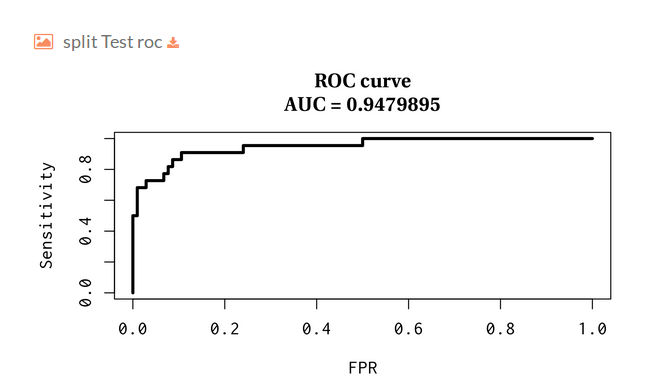
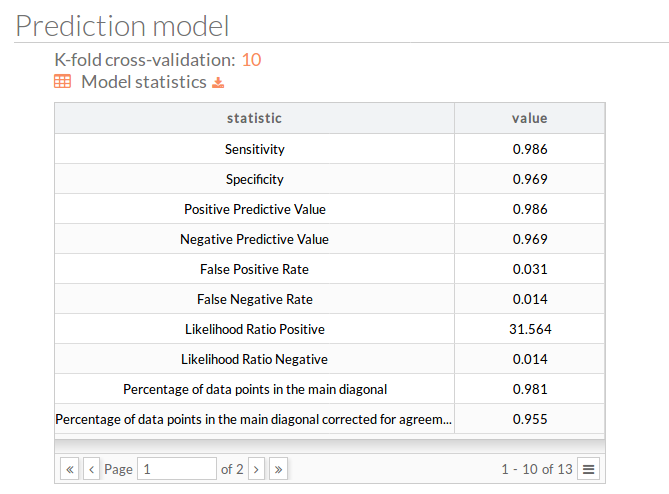
You can download the model statistics.
The prediction tool is based on a machine learning module, this module of the Hipathia web tool can be summarized as follows:
glmnet r package which implements a fast coordinate descent version of the LASSO [5]).C cost or margin and γ) of a non-linear SVM [6] with a radial-basis kernel:C we train a svmC), i.e. the ones with the lower CV mean error.SVM training has been carried out using one of the most powerful libraries to train svm-based models LIBSVM [2] by means of the R interface provided by the package e1071 [1].PRROC [3]The experiment consists in classifying a giving sample as Luminal A or Luminal B (molecular subtype). We use TCGA data (processed by Inma), no pathway filtering was done by hand.
Model Analysis
Hyperparameter search:
CV Performance: CV stats
The most relevant features along with their interaction sign:
| features | coefs |
| :—————————————————————– | :—– |
| p53 signaling pathway: CDK1 CCNB3 | + |
| p53 signaling pathway: CDK2 CCNE1 | + |
| p53 signaling pathway: SERPINB5 | - |
| Oocyte meiosis: REC8* | + |
| Neurotrophin signaling pathway: NFKB1 | + |
| Neurotrophin signaling pathway: RHOA | + |
| Amphetamine addiction: ARC | - |
| RIG-I-like receptor signaling pathway: CHUK IKBKB IKBKG | + |
| Ras signaling pathway: RAP1A | - |
| Progesterone-mediated oocyte maturation: CDK1 | + |
| Vascular smooth muscle contraction: ACTA2 | - |
| Complement and coagulation cascades: BDKRB1 | + |
| Fanconi anemia pathway: RAD51C | + |
| TGF-beta signaling pathway: ROCK1 | - |
| ErbB signaling pathway: ELK1* | - |
| HTLV-I infection: TP53 TBPL2 | + |
| Platelet activation: ITPR1 | - |
| Pathways in cancer: E2F1 | + |
| PI3K-Akt signaling pathway: BCL2 | - |
| Maturity onset diabetes of the young: NKX6-1 | + |
| Signaling pathways regulating pluripotency of stem cells: MAPK1* | - |
| Rap1 signaling pathway: THBS1 | + |
| HTLV-I infection: E2F1 | + |
| Jak-STAT signaling pathway: CDKN1A | + |
| Epstein-Barr virus infection: RB1 | - |
| Colorectal cancer: BIRC5 | + |
| Signaling pathways regulating pluripotency of stem cells: MYC | - |
| Taste transduction: C00076* | - |
| Hepatitis B: JUN | - |
| Axon guidance: GSK3B | - |
| MAPK signaling pathway: MAPT | - |
| cAMP signaling pathway: HHIP | - |
| Fanconi anemia pathway: BRCA1 | + |
| Pathways in cancer: CSF3R | + |
| Cell cycle: CDC45 MCM7 MCM6 MCM5 MCM4 MCM3 MCM2 | + |
| ErbB signaling pathway: CDKN1A | + |
| HTLV-I infection: PTTG2 | + |
| AMPK signaling pathway: CCNA2 | + |
| Oocyte meiosis: CDC25C* | + |
| Non-alcoholic fatty liver disease (NAFLD): BAX | + |
| Hepatitis B: PCNA | + |
| AMPK signaling pathway: G6PC | - |
| Adrenergic signaling in cardiomyocytes: BCL2 | - |
| HTLV-I infection: ANAPC10 CDC20 | - |
| Progesterone-mediated oocyte maturation: CDK1* | + |
| Complement and coagulation cascades: C4A | - |
| Choline metabolism in cancer: WAS | + |
| ErbB signaling pathway: STAT5A | - |
| Herpes simplex infection: FOS | - |
| Amyotrophic lateral sclerosis (ALS): DERL1 | + |
| AGE-RAGE signaling pathway in diabetic complications: F3 | - |
| Non-alcoholic fatty liver disease (NAFLD): PKLR | + |
| Maturity onset diabetes of the young: FOXA3 | - |
| AMPK signaling pathway: CPT1C | + |
| PPAR signaling pathway: FADS2 | + |
| Rap1 signaling pathway: C00076* | + |
| cAMP signaling pathway: GRIN3A | + |
| Glutamatergic synapse: CACNA1A | - |
| Progesterone-mediated oocyte maturation: MAPK14 | - |
| Salivary secretion: BEST2 | + |
| Vibrio cholerae infection: PDIA4 | + |
| cAMP signaling pathway: PLN | + |
| Neurotrophin signaling pathway: JUN | - |
| Pathways in cancer: CCNA1 | - |
| Epithelial cell signaling in Helicobacter pylori infection: GIT1 | - |
| Renal cell carcinoma: TGFA | - |
| Influenza A: RNASEL | + |
| Thyroid hormone signaling pathway: TP53* | + |
| Epithelial cell signaling in Helicobacter pylori infection: CXCL1 | - |
| Signaling pathways regulating pluripotency of stem cells: MAPK14 | + |
| Hepatitis C: EIF2S1 | + |
| Proteoglycans in cancer: CTNNB1 | + |
| Influenza A: STAT1 IRF9 | + |
| Thyroid hormone signaling pathway: CTNNB1 | - |
| Taste transduction: PKD1L3 PKD2L1 | + |
| Prostate cancer: C16038 | - |
| Basal cell carcinoma: PTCH1* | - |
| Toxoplasmosis: C06314 | - |
| Prostate cancer: BCL2 | - |
| Measles: EIF2S1 | + |
| Acute myeloid leukemia: CCNA1 | - |
| Glucagon signaling pathway: CPT1C* | + |
Split Analysis
Split Performance: Test stats
PR curve over the test:
ROC curve over the test set:
Probability for the test set: