This is an old revision of the document!
Worked example : Perturbation effect
In this example, we will use a sample of Pancreatic ductal adenocarcinoma (PDAC) from the repository TCGA -The Cancer Genome Atlas-. In order to simulate the effect of such treatment mentioned in several previous studies [ Blasco MT et al. Complete Regression of Advanced Pancreatic Ductal Adenocarcinomas upon Combined Inhibition of EGFR and C-RAF. ], we will use the erlotinib drug and apply a knock-out of RAF1.
This simulation is carried out by changing the expression level of the genes of interest, in this case are : EGFR and RAF1, recalculating the resulting signaling profile in the simulated condition and comparing it to the original profile.
Similarly, the effect of inhibitory or agonistic drugs can be simulated by decreasing or increasing the expression level of the target gene. This module of analysis brings the functionality of the PathAct algorithm to the HiPathia application. The erlotinib drug is an EGFR inhibitor, that why in this example we will notice that the application of erlotinib drug decreases the expression level of each node that has EGFR in its gene list.
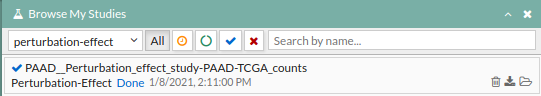
For reproducing this example you can click Perturbation effect example under the help button or you can launch a new study by following these steps:
- Log in Hipathia using login button on the top right corner, the login panel will appear. you can also login using an anonymous user. Please visit Logging in in to further information on this step.
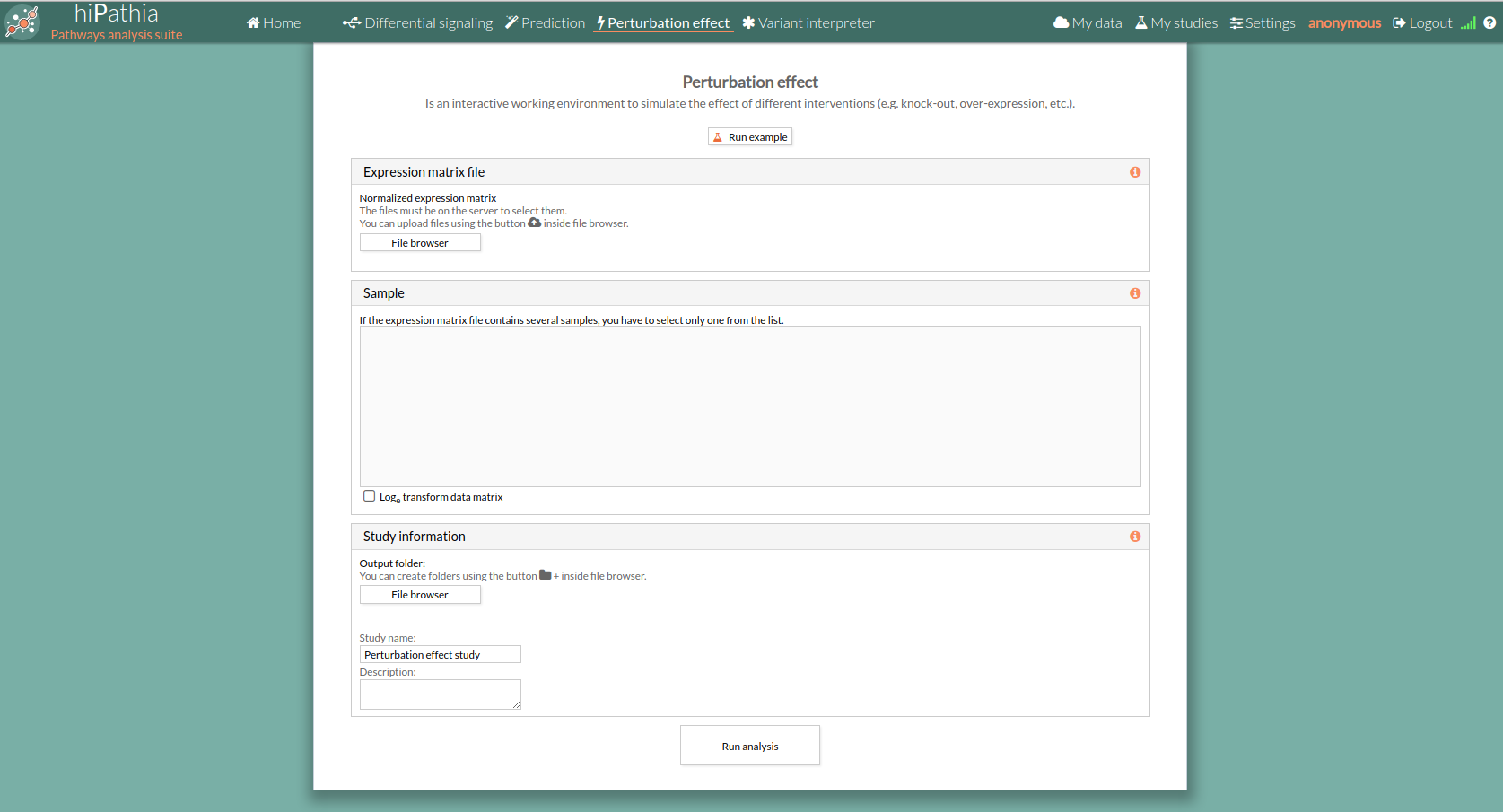
- By clicking the Perturbation effect tool in the menu, the empty form will appear.
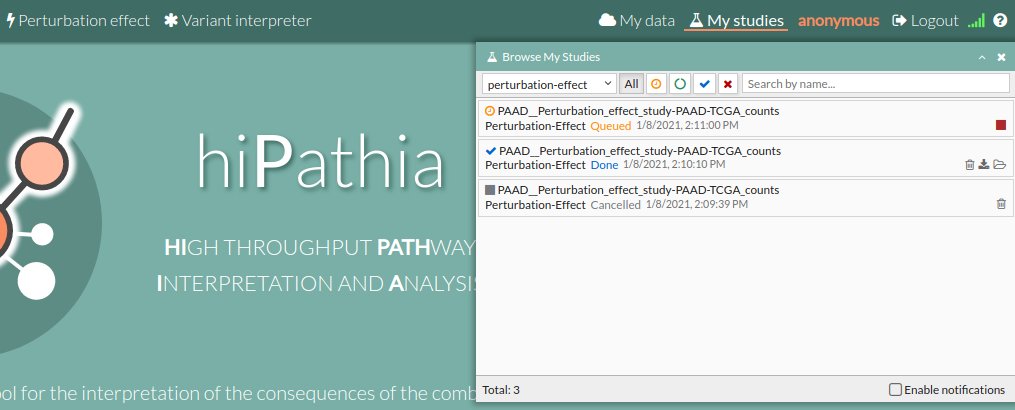
- Press the run example button, A study will appear on the My studies panel and will be processed.
- Once finished, click on it to open the view window.
- The view window will appear.
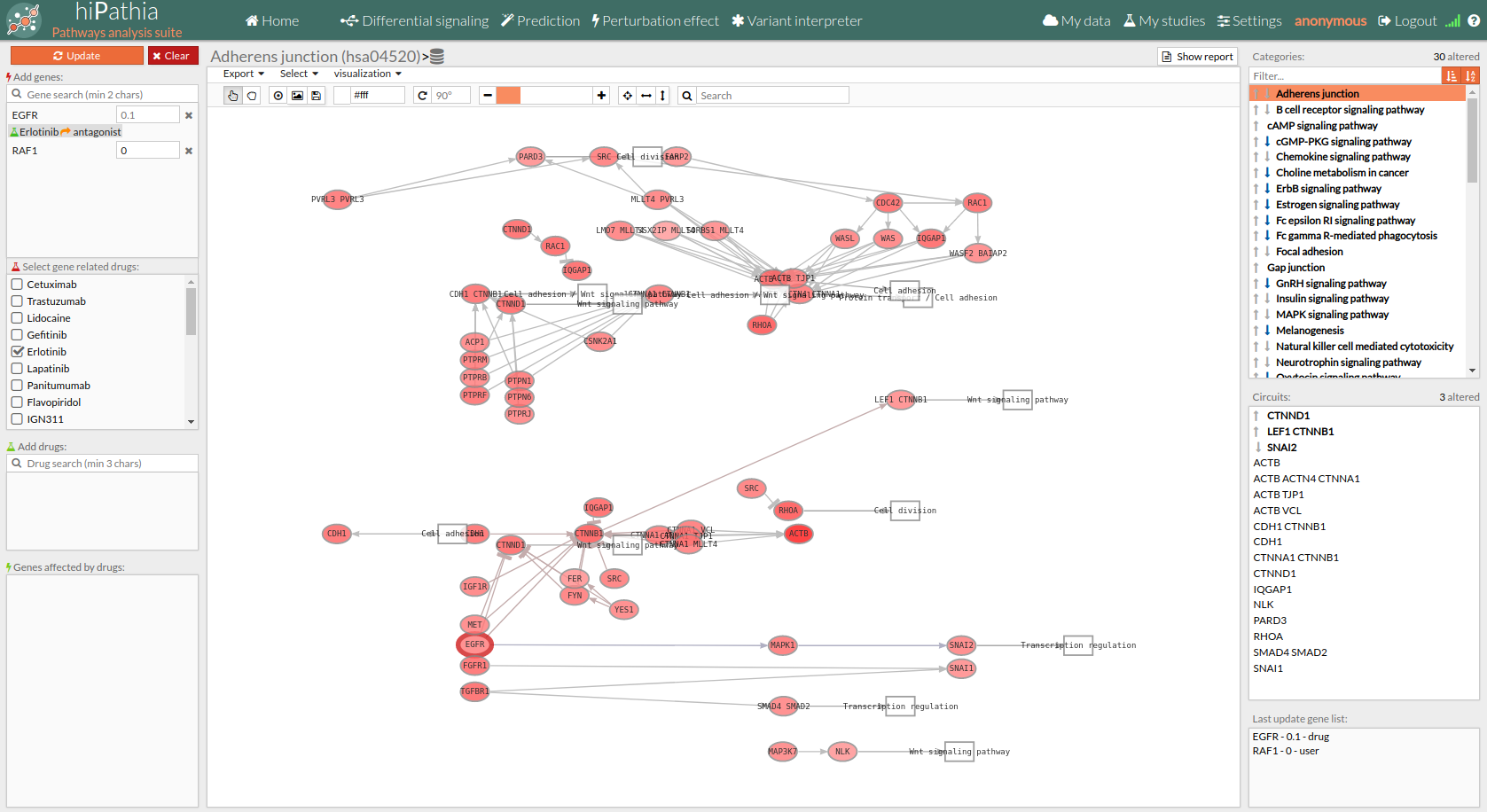
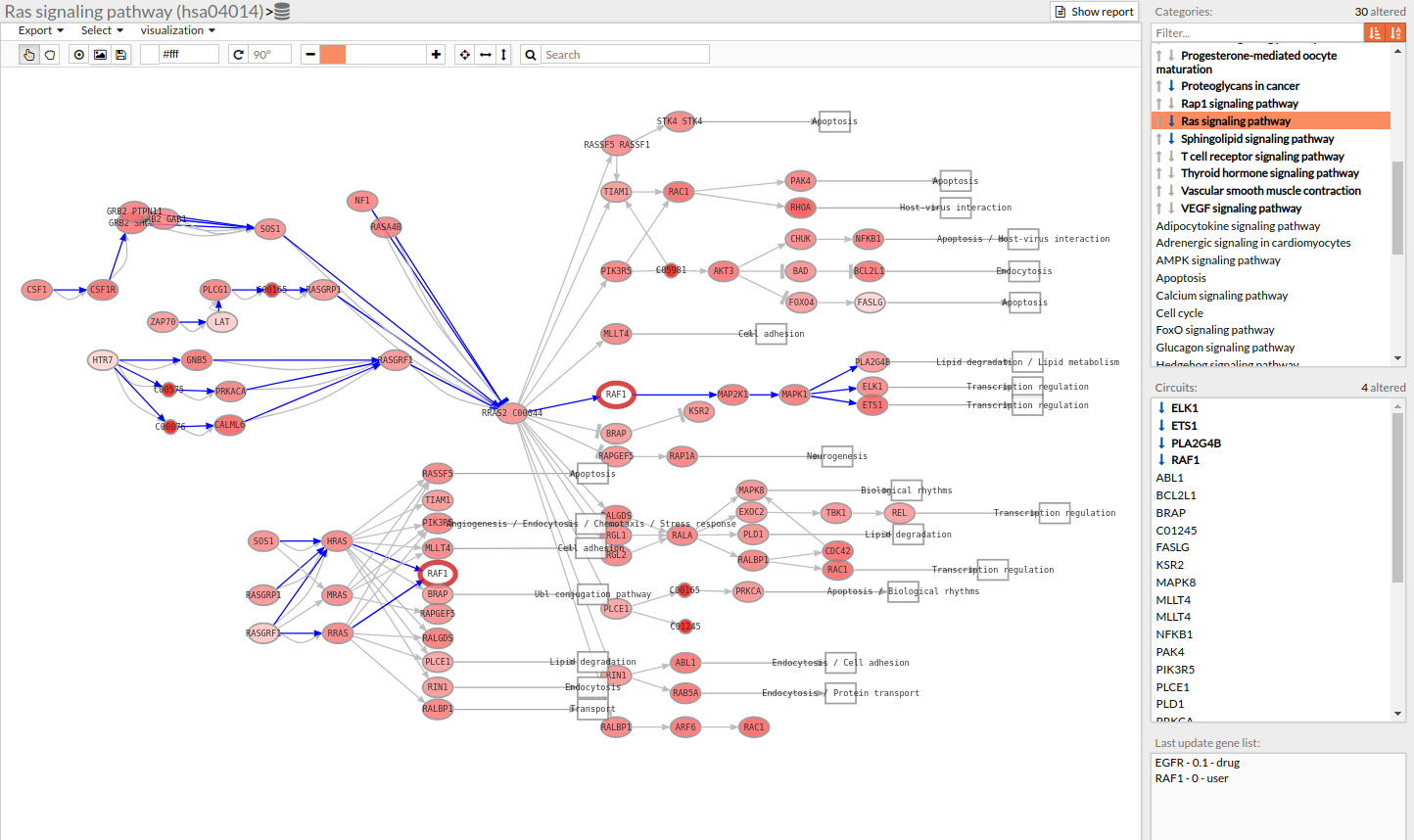
- If you select the genes with a thick red border (as shown in the following image) you will notice that one of its genes is one of our chosen genes targets, and also you can see in this example that the Erlotinib drug (antagonist) has changed the expression value of EGFR to 0 (inhibitory effect). You can either click on a gene on the pathway viewer and change its value.
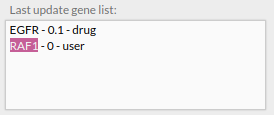
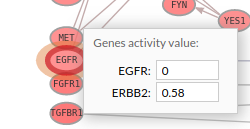 In the add genes panel you can see all targeted genes and their expression levels after such treatment simulation.
In the Search genes box ,of the Add genes panel, you can add more genes and another genes.
In the add genes panel you can see all targeted genes and their expression levels after such treatment simulation.
In the Search genes box ,of the Add genes panel, you can add more genes and another genes.
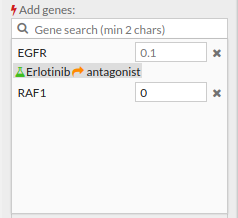
- You can modify the expression level of the genes listed in the Add genes panel and remove them from the list by pressing the X button to their right. The expression level of the removed genes will return to their original value.
Gene perturbations can be studied one at a time or simultaneously in combinations. In this way, it is possible to check for the potential effect of drug combinations over the activity of signaling pathways in the Pancreatic ductal adenocarcinoma sample. For this example we fix the following expression values:
- EGFR (by Erlotinib drug)
- RAF1 (KO)
Resutls and discussion
Perturbation effect tool infers the effect of the knock-out or over-expression of a set of genes on the signaling pathways activity by comparing the signaling values of the pathways in the uploaded sample and the signaling values resulting from the modification of the expression values in the current sample by the selected values.
Each time the Update button on the Gene list panel is pressed, a comparison is performed with the modifications present in the Gene list and the Additional drug targets.
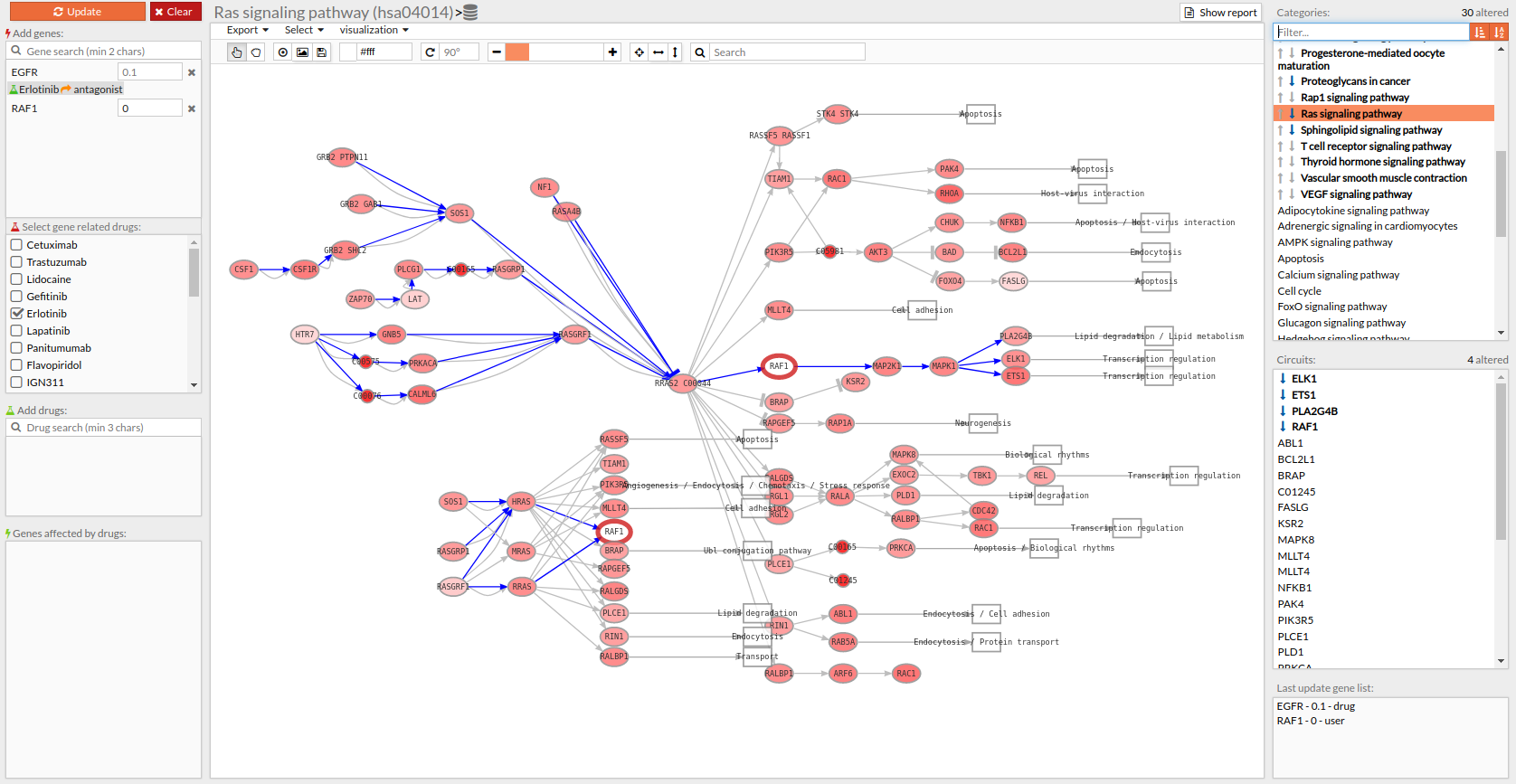
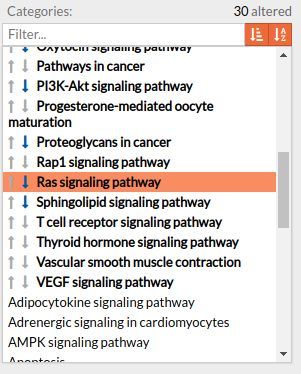
Pathways with signaling differences are depicted in bold characters in the Pathway list panel “Categories panel”. The up and down arrows indicate that there exist up- and down-regulated circuits in the corresponding pathways, respectively. When the up arrow is red or the down arrow is blue, the differences are significant. An absolute fold change greater than 2 is considered significant. Therefore, an absolute log fold change greater than log(2)= 0.6931472 is considered significant.
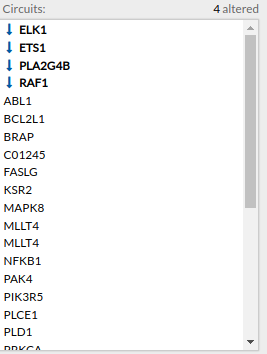
Circuits with signaling differences are depicted in bold characters in the Circuit list panel. The up and down arrows indicate that the circuit is up- or down-regulated, respectively. When the up arrow is red or the down arrow is blue, the differences are significant.
Results can be also visualized in the Pathway viewer in a graphical way. Each time a pathway is selected in the Pathway List it is displayed in the Pathway viewer. The edges of the circuits with a difference in their activation are depicted in different colors.
- Light greyish red: The circuit is up-regulated, but not significantly.
- Light greyish blue: The circuit is down-regulated, but not significantly.
- Red: The circuit is significantly up-regulated.
- Blue: The circuit is significantly down-regulated.
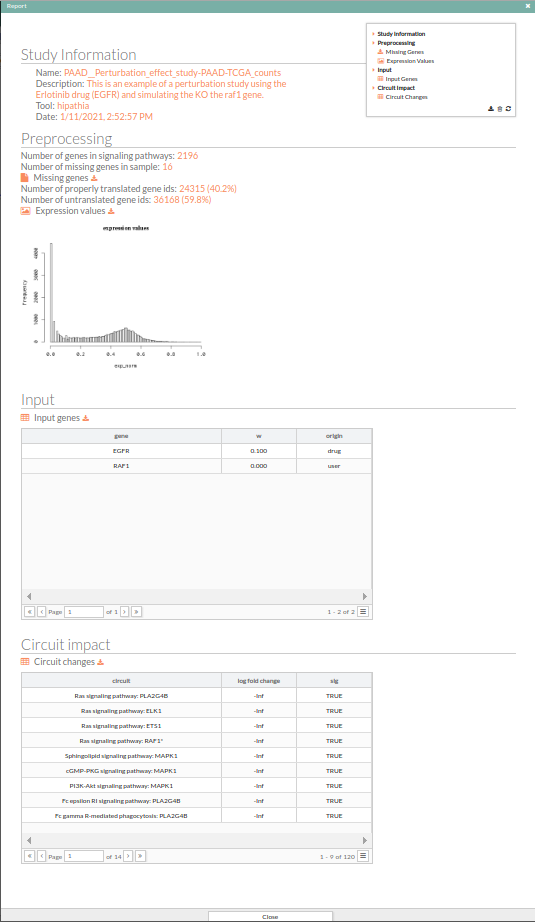
The results can be exhaustively examined and downloaded from the report generated by pressing the Show report button. A new window with the report of the results for the current comparison is displayed. This report includes two fields:
- Input: Lists the genes whose expression has been modified in the current comparison, and the new expression value.
- Path impact: Lists the names of the pathways, ordered by the absolute value of their fold change (also log(Fold_Change)). An absolute fold change greater than 2 is considered significant. Therefore, an absolute log fold change greater than log(2)= 0.6931472 is significant.
In order to close this window and return to the tool page, you can press the close button on the right upper side of the window or the Close button at the bottom of the panel. the report panel is movable.
- Visualize the results. For further information in the interpretation of the results, see Perturbation effect results.