This is an old revision of the document!
Table of Contents
Worked example Prediction
Test inputs
1. Log in into HiPathia. For further information on this step visit Logging in.
2. We will test the model with another Breast Cancer dataset (Luminal A Vs Luminal B) from the repository The Cancer Genome Atlas. You can download the expression matrix of the test data from the link:
- Test expression matrix: brca_genes_vals_bn_test.txt
3. Upload the data to HiPathia in the data panel by clicking on My data. For further information on this step visit Upload your data.
4. Click Prediction button.
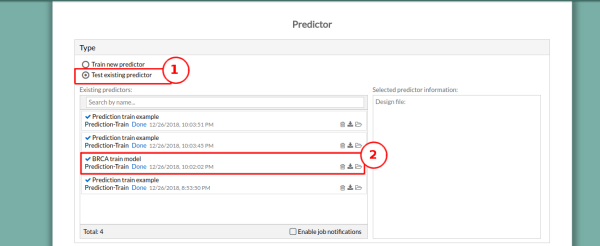
5. In the Type panel, select Test existing predictor. A window with all the existing models will appear. Select the model you want to use. We will use the model we have created in Worked example Prediction - Train. The model information will appear on the right panel.
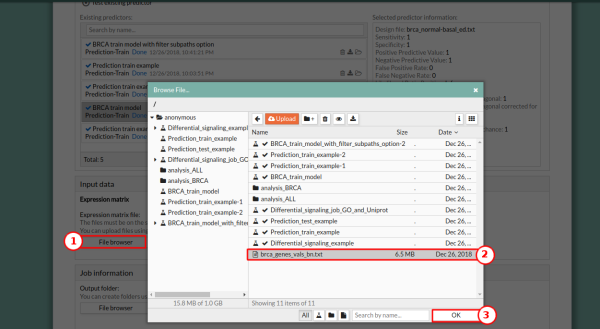
6. In the Input data panel select Expression matrix. Press the File browser of the Expression matrix file, and select the desired file, brca_genes_vals_bn_test.txt.
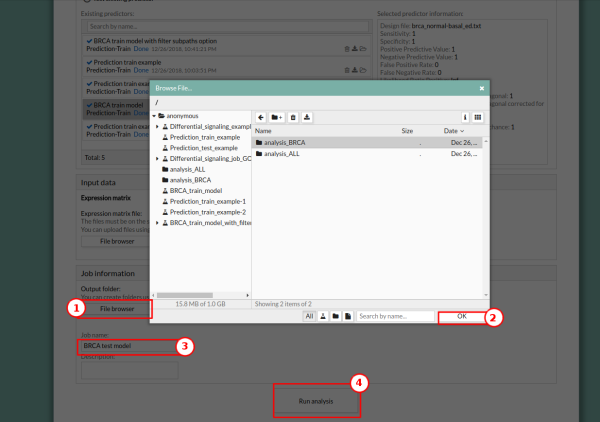
7. In the Job information panel, press the File browser button and select the desired output folder. In this case, we will use analysis_BRCA. Give a name to the study, for example, “BRCA test model”.
8. Press the Run analysis button. A study will be created and listed in the studies panel. You can access this panel by clicking on the My studies button.
Test report
As we said before, this experiment consists in test the trained model (in this example), using another split of data (Luminal A or Luminal B).
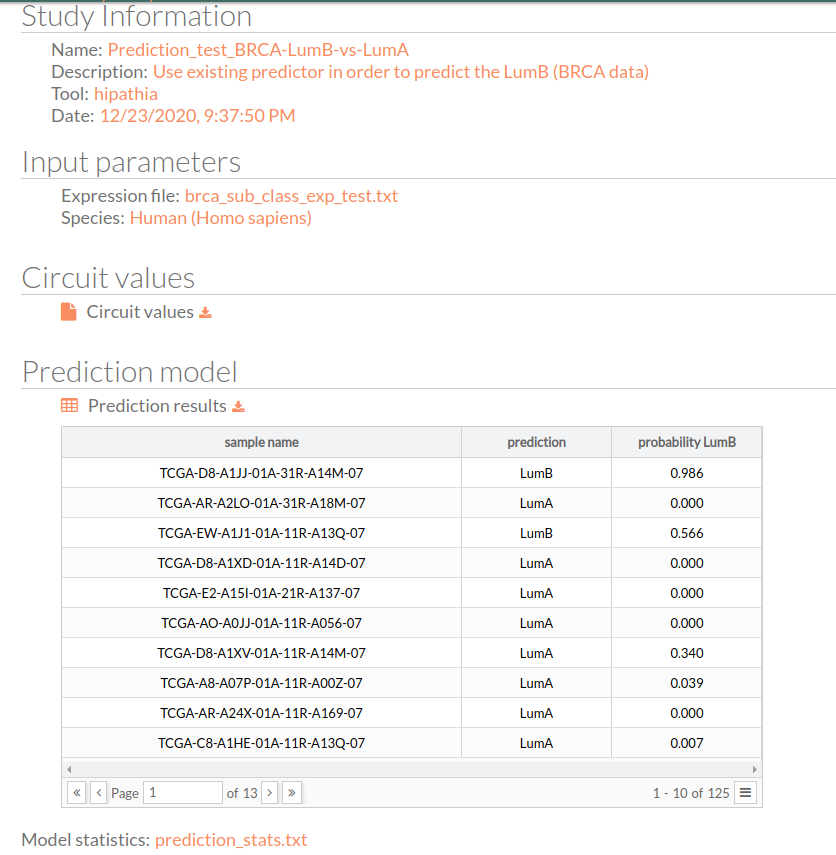
Study Information
Here you can find the information about the selected study.
- Name: the study name.
- Description: the description of the current study.
- Tool: the name of the used tool (in this case, is Hipathia).
- Date: study's launching date (MM/DD/AAAA, HH:MM:SS AM/PM format)
Input Parameters
Here you can find the parameters with which the current study was launched.

- Expression file: The name of the expression file that has been used in the current study.
- Species: The species of this experiment; Human (Homo sapiens),Mouse (Mus musculus) or Rat (Rattus norvegicus).
Circuit values
You can download the matrix of circuit activity values by clicking on circuit values. This matrix file indicates for each “effector circuit” the level of activation calculated using Hipathia method for each sample.
Prediction model
This is the most important result of our predictor, which is a matrix with three columns :
- Sample name: all the 125 samples in the used expression matrix file.
- Prediction: the predicted group LumB (Luminal B) or LumA (Luminal A)
- Probability LumB: this is the probability of being lumB, if it is 1 that means the predictor is 100% sure that the given result will be LumB.
You can download the matrix of predicted experimental design by clicking on Prediction results.
Prediction evaluation
Confusion Matrix and Statistics
Reference
Prediction LumA LumB
LumA 95 5
LumB 9 16
Accuracy : 0.888
95% CI : (0.8192, 0.9374)
No Information Rate : 0.832
P-Value [Acc > NIR] : 0.0547
Kappa : 0.6277
Mcnemar's Test P-Value : 0.4227
Sensitivity : 0.9135
Specificity : 0.7619
Pos Pred Value : 0.9500
Neg Pred Value : 0.6400
Prevalence : 0.8320
Detection Rate : 0.7600
Detection Prevalence : 0.8000
Balanced Accuracy : 0.8377